2019
Machine learning and data analytics platform for infectious disease genetics. — Our group’s focus is to foster research in data and computation intensive research areas. The last two decades have seen an unprecedented change in almost all areas of sciences. Before that most disciplines were determined by the scarcity of experimental data. The exponential pace of microelectronics development has changed this, on one hand by making available high throughput sensors and digital instruments and on the other by providing high speed computers with large storage and fast interconnecting network. Beyond the almost limitless opportunities there are demanding challenges, too: how to handle the data avalanche from experiments, how to get out the most from information technology in various scientific disciplines, and how to understand and manages the ever-growing complexity of the computational system itself. We study computer networks and systems like it was a “natural phenomena” and with continuously following the technologies, we use them for analyzing science data in various fields from genomics to cosmology.
We are part of a large European H2020 project, COMPARE in which bioinformatics tools are developed for outbreak detection. The health of humans and animals around the world is increasingly under threat due to new and recurring epidemics and foodborne disease outbreaks, which place pressure on health services and the production of livestock. These epidemics also reduce consumer confidence in food and negatively impact trade and food security. The longer it takes from the start of an outbreak of for example Ebola, influenza or salmonella until it is detected and stopped, the greater the consequences. The most important factor in being able to limit the consequences and costs of such outbreaks is the ability to quickly identify the disease-causing microorganisms that are causing the disease. Also, there is the need for knowledge about the mechanisms that cause the disease, and how the bacteria are transmitted to and between humans. The goal of the COMPARE project is a better surveillance system for infectious diseases, to speed up the detection of and response to disease outbreaks among humans and animals worldwide using new genome technology. Our group is responsible for the advanced database and data analysis system which will store, analyse and share the genomic data collected by researchers all over the world. We develop a “virtual research environment”, where interested partners can log in, and use the already installed tools, software and data together with their own to do research (Fig. 1). Wigner Cloud is used as a hardware backend for developing the portal. We are also involved in the development of machine learning methods, like artificial neural networks for inferring antibiotic resistance based on the genetic sequences of bacteria.
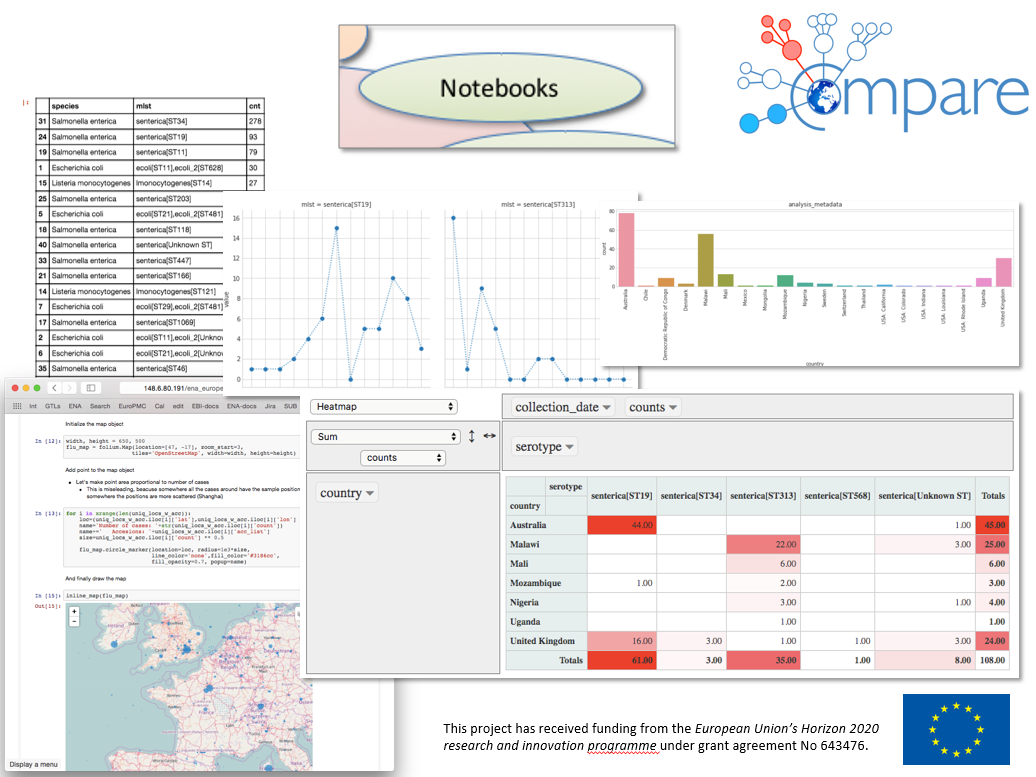
Figure 1. Snapshot of pathogen genome data analysis in the COMPARE Data Hub.



